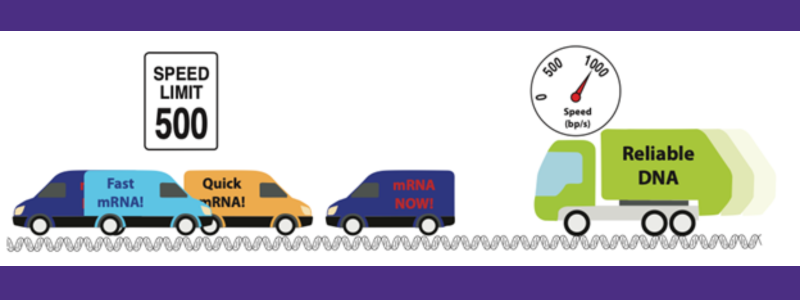
An analogy between replication dynamics and neighborhood traffic. A new approach measures the replication pauses caused by active RNA transcription. Courtesy of Paul Wiggins.
Investigators in the physics, bioengineering, and microbiology departments at the University of Washington have developed a novel method for measuring the speed of polymerase enzymes on different regions of the genome inside living cells. It turns out, much like driving a car, the speed enzymes move along the DNA depends on where they are. There are short pauses (think stop signs) as well as longer periods of fast and slow speed (think different speed limits).
Why does the speed of the enzyme change? One known cause is a phenomenon called replication conflicts. The researchers in the Wiggins lab compared the enzyme motion to traffic: Think of the genome as a narrow road where many delivery trucks are moving constantly back and forth between houses (this is transcription, the first step in the process in which proteins are made) at the same time a recycling truck is trying to make a single trip to every house in order (this is the replication process in which DNA is copied). The trucks cannot pass each other easily and this is essentially the mechanism for conflicts between the replication and transcription processes.
The Wiggins lab and their collaborators in the Vanderbilt School of Medicine Merrikh lab had previously used single-molecule fluorescence microscopy to watch these conflicts occur. “Our old approach had the time resolution to observe the conflicts; however, we never knew where these conflicts were occurring on the genome,” said Dean Huang, a graduate student in the Department of Physics and the first author on the study. In the new study, published in the journal Nature Communications, the researchers describe a novel sequencing-based approach to characterize the motion. “This approach has a huge advantage since we know exactly where the replication machinery stops, slows down or speeds up and we can measure the speed everywhere at the same time,” said Paul Wiggins, joint associate professor of physics, bioengineering and microbiology and the lead author of the study. “Before this approach was developed, it was as if we knew trucks were stopping but not where. Now we are able to observe that the truck is stopping at specific locations, and this allows us to investigate the cause of the slow downs.”
This approach has a huge advantage since we know exactly where the replication machinery stops, slows down or speeds up and we can measure the speed everywhere at the same time. – Paul Wiggins
As expected, the researchers observed long pauses (minutes) at severe engineered conflicts between transcription and replication, but even in unmodified cells, short pauses on the order of seconds are observed at the most active genes. The new approach allows the duration of these pauses to be measured for the first time. “We believe this new approach will be a critical tool for understanding how the cells resolve conflicts between biological processes that compete over the same DNA,” Huang said. “If the cell cannot manage these conflicts, they can lead to DNA damage and cell death or mutations, which in turn can lead to antibiotic resistance in bacteria or cancer in our own cells.”
These pauses may not be the most novel feature in the replication data. The scientists were also able to measure the speed of the replication complex over different regions of the genome for the first time and they discovered unexpected features in this data that are not explained by conflicts. The measurements reveal evidence for several potential factors that affect the speed. “To our surprise, we observe long genomic-scale oscillations in the speed. At first, we believed that these were fast and slow regions of the genome,” said Wiggins. “These are a bit like parts of a road with faster and slower speed limits. However, by perturbing the cells, we quickly learned that the speed was not determined by position, but rather by timing.”
We believe this new approach will be a critical tool for understanding how the cells resolve conflicts between biological processes that compete over the same DNA. If the cell cannot manage these conflicts, they can lead to DNA damage and cell death or mutations, which in turn can lead to antibiotic resistance in bacteria or cancer in our own cells. – Dean Huang
Speeds oscillate in time relative to the start of the replication process and two replication complexes have roughly the same speeds at the same time, even though they are replicating different locations on the genome. Why does the speed oscillate? “We don’t yet know for sure, but there are a couple of exciting possibilities,” says Wiggins. Learn more about the research and these possibilities in the paper.


