Professor
hsauro@uw.edu
Phone: (206) 880-8093
Office: Foege N410A
Herbert Sauro
We are developing the standards and best practices for developing predictive models of biological systems and disease states
Creating software to enhance advanced modeling
Developing whole cell metabolic models
Developing models of protein signaling networks related to cancer tumor growth
Understand the design of protein signaling networks in terms of signal processing
Developing best practices for models that may be used in a clinical settings
1. Use mathematics and computation to help understand the dynamics and operation of cellular processes.
2. Develop new more reliable approaches to modeling complex systems such that the models can be used with confidence in clinical settings.
3. Disseminate the work to the wider scientific community and support best practices in systems biology modeling multi-scale modeling of subcellular and multicellular systems.
MSc: Biological Computation, University of York
PhD: Systems Biology, Oxford brookes, UK
PGCE: Teaching Degree, University of Aberystwyth
BIOEN 499: Special Topics: Modeling in Systems Biology
Entus, R, B Aufderheide, and HM. Sauro. “Design and implementation of three incoherent feed-forward motif based biological concentration sensors.” Systems and synthetic biology 1, no. 3 (2007): 119-128.
Galdzicki, M, KP. Clancy, E Oberortner, M Pocock, JY. Quinn, CA. Rodriguez, N Roehner et al. “The Synthetic Biology Open Language (SBOL) provides a community standard for communicating designs in synthetic biology.” Nature biotechnology 32, no. 6 (2014): 545.
Fell, DA., and HM. Sauro. “Metabolic control and its analysis.” The FEBS Journal 148, no. 3 (1985): 555-561.
Kim, KH., and HM. Sauro. “Adjusting phenotypes by noise control.” PLoS computational biology 8, no. 1 (2012): e1002344.
Kim, KH, H Qian, and HM. Sauro. “Nonlinear biochemical signal processing via noise propagation.” The Journal of chemical physics 139, no. 14 (2013): 10B608_1.
Sleight, SC., BA. Bartley, JA. Lieviant, and HM. Sauro. “In-Fusion BioBrick assembly and re-engineering.” Nucleic acids research 38, no. 8 (2010): 2624-2636.
Paladugu, SR., V. Chickarmane, A. Deckard, J. P. Frumkin, M. McCormack, and H. M. Sauro. “In silico evolution of functional modules in biochemical networks.” IEE Proceedings-Systems Biology 153, no. 4 (2006): 223-235.
Sauro, HM. Enzyme kinetics for systems biology. 2nd Edition, Future Skill Software, 2012.
Sauro, HM. Systems Biology: Introduction to Pathway Modeling, Future Skill Software, 2012.
Hucka, M, A Finney, HM. Sauro, H Bolouri, JC. Doyle, H Kitano, AP. Arkin et al. “The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models.” Bioinformatics 19, no. 4 (2003): 524-531.
Medley, M, Choi K, Matthias K, Smith L, Gu S, Hellerstein J, Sealfon SC, Sauro HM Tellurium Notebooks – An Environment for Dynamical Model Development, Reproducibility, and Reuse (2018), PLoS Comp Bio,.
In the News
UW Bioengineering faculty embrace AI tools to enhance teaching and learning
2025-03-21T07:09:14-07:00March 18th, 2025|
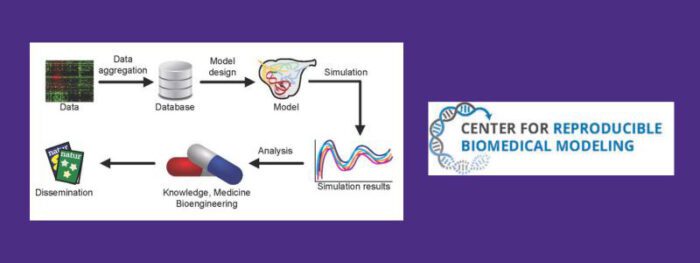
NIH provides major support to CRBM for advancing reproducible biomedical modeling
2024-06-10T11:56:24-07:00June 10th, 2024|
UW Bioengineers pivot to develop coronavirus solutions
2022-08-01T14:43:01-07:00July 9th, 2020|
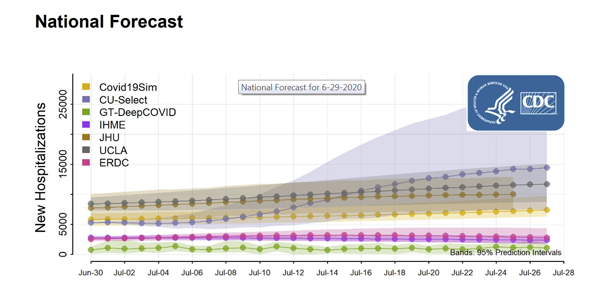
How Credible are the COVID-19 Models? Center Aims to Find Out
2022-08-04T03:36:13-07:00July 7th, 2020|
Herbert Sauro, David Baker, Dayong Gao Elected to AIMBE
2020-10-26T08:11:46-07:00May 27th, 2020|
Jay Rubinstein Inducted as 2019 AIMBE Fellow
2020-10-26T08:11:59-07:00May 23rd, 2019|